运行结束后输出密度rho
-
@wangfei9088 请教大佬,参照specieReactionRates的写法,先尝试输出每个反应步的体积平均Qdot(命名sumVQdotRi/V),计算Qdot时需要物种的比焓h[speciei],这个如何访问呢?
const scalar h[speciei] = specieThermos_h[speciei].hf(); //如何访问?
bool Foam::functionObjects::reactionSpecieQdots::write() { logFiles::write(); const label nSpecie = chemistryModel_.nSpecie(); const label nReaction = chemistryModel_.nReaction(); // Region volume const scalar V = this->V(); for (label reactioni=0; reactioni<nReaction; reactioni++) { if (Pstream::master()) { writeTime(file()); file() << token::TAB << reactioni; } const PtrList<volScalarField::Internal> RR ( chemistryModel_.reactionRR(reactioni) ); scalar sumVQdotsR = 0; for (label speciei=0; speciei<nSpecie; speciei++) { scalar sumVQdotRi = 0; const scalar h[speciei] = specieThermos_h[speciei].hf(); //如何访问? if (all()) { sumVQdotRi = fvc::domainIntegrate(RR[speciei]*(-h[speciei])).value(); } else { sumVQdotRi = gSum ( scalarField ( fvMeshFunctionObject::mesh_.V()*RR[speciei]*(-h[speciei]), cells() ) ); } if (Pstream::master()) { file() << token::TAB << sumVQdotRi/V; } sumVQdotsR += sumVQdotRi; } if (Pstream::master()) { file() << token::TAB << sumVQdotsR/V; } if (Pstream::master()) { file() << nl; } } if (Pstream::master()) { file() << nl << endl; } return true; } -
@wangfei9088 这是我看到基金会版本里面用来算Qdot的,我直接贴上去了。我的问题是specieThermo_这个可以直接访问吗?const label nSpecie = chemistryModel_.nSpecie();可以这么写是前面定义了
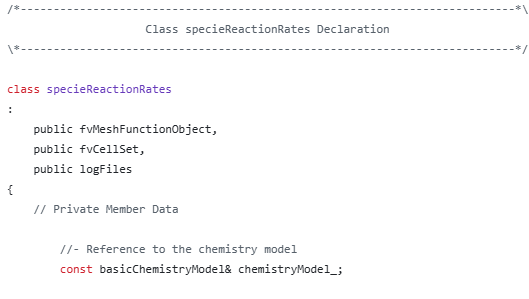
我看到specieThermo_是在别的地方定义的。我弄不明白这个。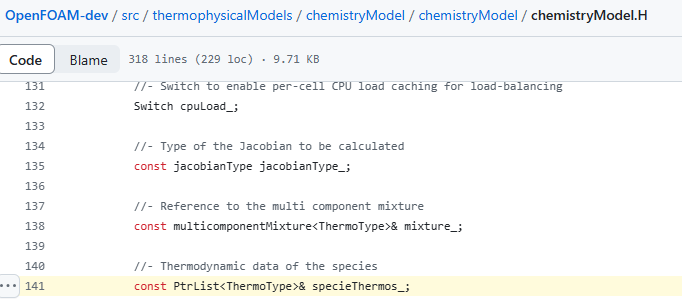
-
@尚善若水 看了一下,开发版和of2012差别很大,开发版里不能用specieThermo_了。依据要不要单位,组分的生成焓在开发版里可以这么写:
dimensionedScalar hi = chemistryModel_.thermo().hfi(speciei); //- Enthalpy of formation [J/kg] 或者 scalar hi = chemistryModel_.thermo().hfiValue(speciei); //- Enthalpy of formation [J/kg] -
@wangfei9088 好的,我仔细看下。谢谢。
-
@wangfei9088 感谢大佬,程序正常输出,正确性还没测试,也许可以和cantera的结果比一下,代码就是上面发的加上根据大佬说的改的。另外,我在想可不可以更进一步,写出每一个反应步每个组分的Qdot,命名为Qdot.Ri.Speciei,类似输出RR.Speciei?
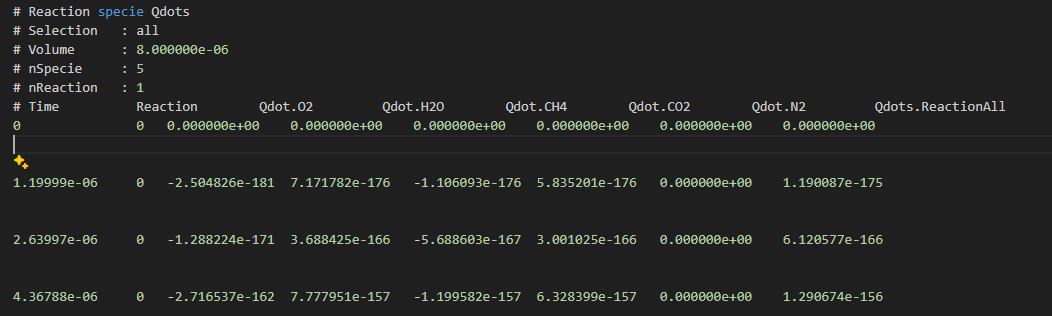
-
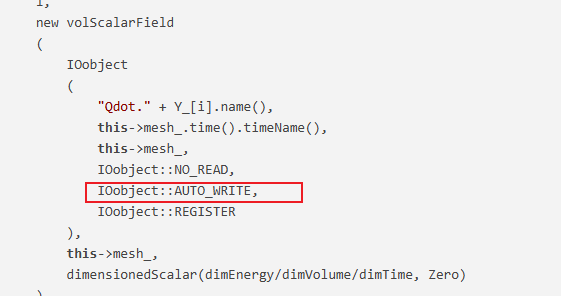
对于你的问题,我最近也在修改,加入以下代码,然后在类定义中添加成员变量就行了。我这边是可以成功输出每个物质对应的Qdot
template<class ReactionThermo, class ThermoType> Foam::tmp<Foam::volScalarField> Foam::StandardChemistryModel<ReactionThermo, ThermoType>::Qdot() const { tmp<volScalarField> tQdot ( new volScalarField ( IOobject ( "Qdot", this->mesh_.time().timeName(), this->mesh_, IOobject::NO_READ, IOobject::NO_WRITE, IOobject::NO_REGISTER ), this->mesh_, dimensionedScalar(dimEnergy/dimVolume/dimTime, Zero) ) ); QdotSpecies_; if (!QdotSpecies_.size()) { // 首次调用时初始化 QdotSpecies_.setSize(nSpecie_); forAll(Y_, i) { QdotSpecies_.set ( i, new volScalarField ( IOobject ( "Qdot." + Y_[i].name(), this->mesh_.time().timeName(), this->mesh_, IOobject::NO_READ, IOobject::AUTO_WRITE, IOobject::REGISTER ), this->mesh_, dimensionedScalar(dimEnergy/dimVolume/dimTime, Zero) ) ); } } if (this->chemistry_) { scalarField& Qdot = tQdot.ref(); Qdot = 0.0; forAll(Y_, i) { volScalarField& QdotI = QdotSpecies_[i]; const scalar hi = specieThermo_[i].Hc(); forAll(Qdot, celli) { QdotI[celli] = -hi*RR_[i][celli]; Qdot[celli] += QdotI[celli]; } // 修正边界条件 QdotSpecies_[i].correctBoundaryConditions(); } tQdot.ref().correctBoundaryConditions(); } return tQdot; }